Bioinformatics
Bioinformatics is an interdisciplinary field that develops and improves upon methods for storing, retrieving, organizing and analyzing biological data.
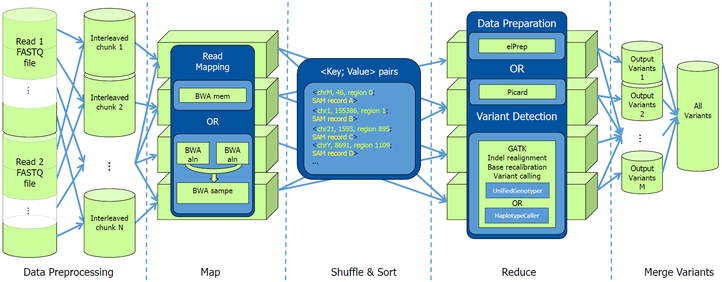
The recent revolution in technology enables the production of molecular data at unprecedented rates. All this raw data needs to be stored, moved and processed, which is a non-trivial task, given the vast volumes of data. The development of software implementations that can extract useful biological knowledge from these data is a challenge, given that most problems are underdetermined and thus require combining the proper algorithms with the right biological assumptions.
Bioinformatics combines biological knowledge with many areas of computer science, mathematics and engineering to process and biological data. We combine complex machines and the use of parallelization and distributed computing to read in and process biological data at a much faster rate than before. Relying on the latest developments in algorithm design, machine learning, artificial intelligence, soft computing, data mining, image processing, and simulation, we develop dedicated solutions for problems in the biomedical, agrotech and microbial sector. We specifically focus on developing high performance solutions for next-generation sequence analysis and for the integrative analysis of genotype-phenotype data.
Staff
Kathleen Marchal, Jan Fostier, Tom Dhaene, Mario Pickavet, Tijl De Bie, Pieter Audenaert, Lieven Verbeke, Dries De Maeyer, Paolo Simeone, Jefrey Lijffijt
Researchers
Camilo Andres, Dieter De Witte, Dries Decap, Mahdi Heydari, Ahmad Mel, Ine Melckenbeek, Giles Miclotte, Mushthofa Mushthofa, Joeri Ruyssinck, Sofie Van Gassen, Robin Vandaele, Bram Weytjens.
Projects
- ICON GAP: Genome Analytics Platform
- IWT-SBO NEMOA: Network-based approaches for the identification and mode of action determination of anti-bacterial agents
- FWO project: De novo genome assembly using both second and third generation sequencing data
- FWO project: Network-based analysis of genotype-expression phenotype data
- FWO project: Genetic adaptation of bacterial catabolic pathways for pesticide biodegradation: role of historical contingency
- Odysseus Grant “Exploring Data: Theoretical Foundations and Applications to Web, Multimedia, and Omics Data”.
Key publications
- Giles Miclotte, Mahdi Heydari, Piet Demeester, Stephane Rombauts, Yves Van de Peer, Pieter Audenaert, and Jan Fostier. 2016. “Jabba: Hybrid Error Correction for Long Sequencing Reads.” Algorithms for Molecular Biology 11: 10.
- Dries Decap, Joke Reumers, Charlotte Herzeel, Pascal Costanza and Jan Fostier, “Halvade: scalable sequence analysis with MapReduce”, Bioinformatics 2015.
- Arne De Coninck, Bernard De Baets, Drosos Kourounis, Fabio Verbosio, Olaf Schenk, Steven Maenhout and Jan Fostier, “Needles: toward large-scale genomic prediction with marker-by-environment interaction”, Genetics 2016.
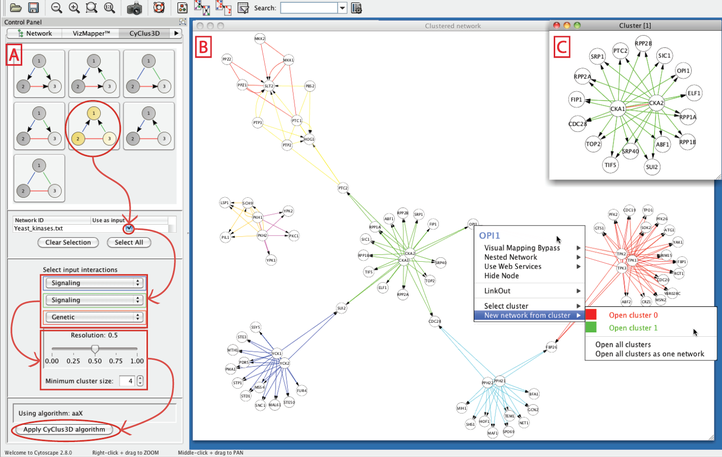
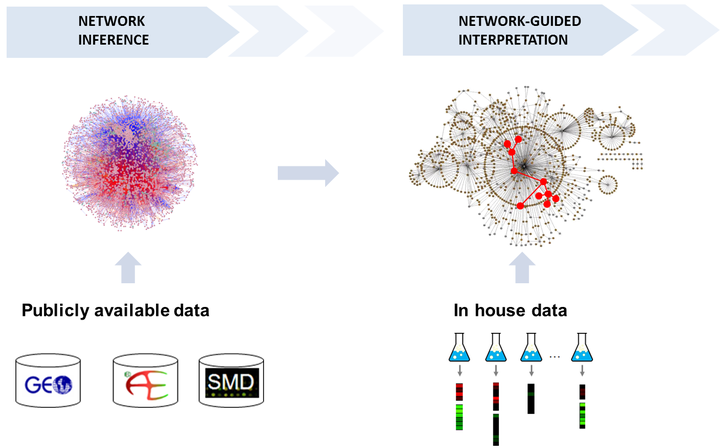
- Lieven Verbeke, Jimmy Van den Eynden, Ana Carolina Elisa Fierro Gutierrez, Piet Demeester, Jan Fostier and Kathleen Marchal UGent. Pathway relevance ranking for tumor samples through network-based data integration. (2015) PLOS ONE. 10(7).
- Dries De Maeyer, Bram Weytjens, Joris Renkens, Luc De Raedt and Kathleen Marchal. (2015). PheNetic: network-based interpretation of molecular profiling data. Nucleic Acids Research 43(W1). p.W244-W250
- Ine Melckenbeeck, Pieter Audenaert, Tom Michoel, Didier Colle and Mario Pickavet, "An algorithm to automatically generate the combinatorial orbit counting equations", Plos ONE 2016.
- Maarten Houbraken, Sofie Demeyer, Dimitri Staessens, Pieter Audenaert, Didier Colle and Mario Pickavet UGent, "Fault tolerant network design inspired by Physarum polycephalum", Natural Computing, 2013.
- Sofie Van Gassen, Britt Callebaut, Mary van Helden, Bart Lambrecht, Piet Demeester, Tom Dhaene and Yvan Saeys, “FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data”, Cytometry Part A, 2015.
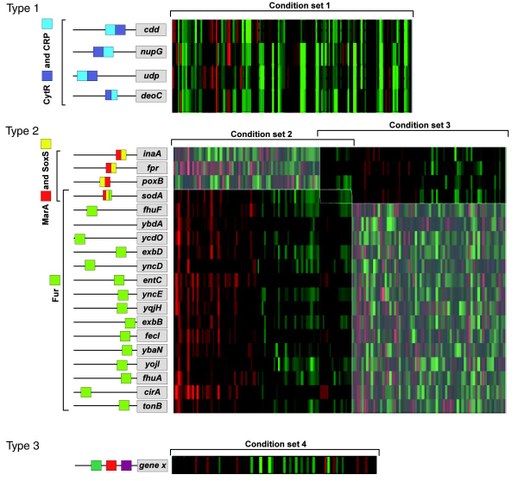
- Lemmens, Karen, Tijl De Bie, Thomas Dhollander, Pieter Monsieurs, Bart De Moor, Julio Collado-Vides, Kristof Engelen, and Kathleen Marchal. 2009. “The Condition-dependent Transcriptional Network in Escherichia Coli.” Ed. G Stolovitzky, P Kahlem, and A Califano. Annals of the New York Academy of Sciences 1158: 29–35.
- De Bie, Tijl, Leon-Charles Tranchevent, Liesbeth MM Van Oeffelen, and Yves Moreau. 2007. “Kernel-based Data Fusion for Gene Prioritization.” Bioinformatics 23 (13): I125–I132.
- Lemmens, Karen, Tijl De Bie, Thomas Dhollander, Sigrid CJ De Keersmaecker, Inge M Thijs, Geert Schoofs, Ami De Weerdt, et al. 2009. “DISTILLER: a Data Integration Framework to Reveal Condition Dependency of Complex Regulons in Escherichia Coli.” Genome Biology 10 (3).
- Lemmens, Karen, Thomas Dhollander, Tijl De Bie, Pieter Monsieurs, Kristof Engelen, Bart Smets, Joris Winderickx, Bart De Moor, and Kathleen Marchal. 2006. “Inferring Transcriptional Modules from ChIP-chip, Motif and Microarray Data.” Genome Biology 7 (5).