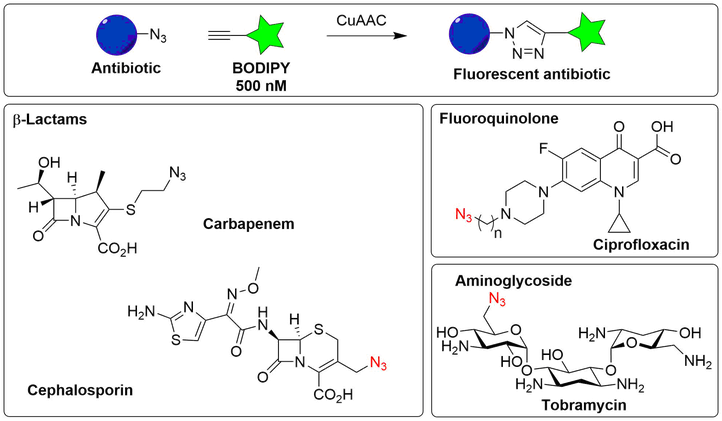
Fluorescently labeled antibiotics for investigating phenotypic properties of mutant bacteria
In a collaborative project with Prof. Tom Coenye, a large collections of evolved mutants with reduced susceptibility will be generated applying an experimental evolution approach. Our group will synthesize fluorescently labelled antibiotics to compare the mutants’ antibiotic uptake and efflux with that of WT strains.
The synthesis of fluorescently-labelled antibiotics has been reported in literature, but such molecules are not commercially available and often suffer from the drawback that they lose (part) of their antimicrobial activity. To obtain labelled antibiotics, we will prepare azide-modified analogues of various classes of antimicrobials commencing from the parent agent or a commercially available precursor thereof. The azide moiety will allow conjugation by means of CuAAC methodology to BODIPY-fluorophores that are equipped with a terminal alkyne.
General overview of the labelling reaction (‘click chemistry’) (upper panel) and examples of labeled antibiotics that will be synthesized (lower panels).
The azide-modified antibiotics are designed to allow introduction of the fluorophore as the final synthetic step under mild conditions, whilst paying close attention to the known structure-activity relationship (SAR) of the compound as not to compromise the antibacterial activity.
For the majority of the classes of antibiotics from our panel, the commercially available agents do not allow direct azide introduction without compromising the SAR, although some can be functionalized in an indirect fashion. Hence, most derivatives have to be prepared from their synthetic precursors (simple or advanced) or can only be accessed through extensive protective group manipulations.