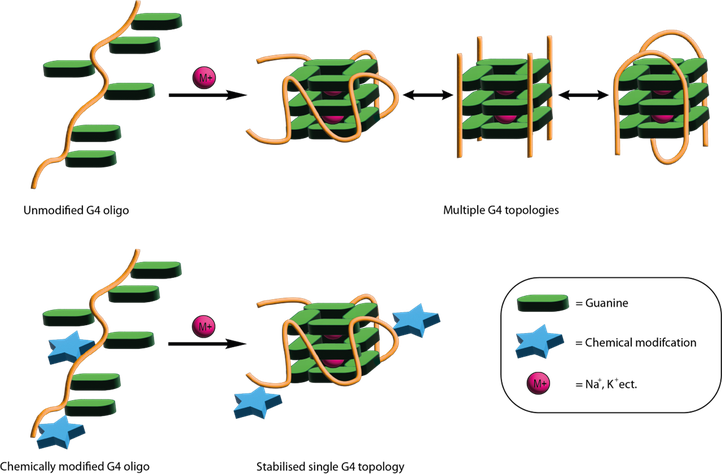Jack Barr
CV
Jack Barr obtained his MChem in Chemistry with International Experience in 2019 from the University of Manchester, United Kingdom. After graduation, Jack was selected for the AstraZeneca R&D Graduate Program located at the AZ Gothenburg site, Sweden where he contributed to a variety of company projects and academic collaborations from 2019 to 2021.
Currently he is pursuing an organic chemistry PhD in the OBCR group under the supervision of Prof. Annemieke Madder within the Marie Curie Innovative Training Network (ITN) OLIGOMED.
His research focuses upon the stabilisation of G-quadruplex forming aptamers, in particular making use of chemical modification methodologies developed within the OBCR group. Key areas of work include phosphoramidite building block synthesis, oligonucleotide synthesis and g-quadruplex characterisation.
Research Project
G-Quadruplex oligonucleotide decoy stabilisation.
Since the discovery that they are numerous in several oncogene promotor regions and play key role in their regulation there has been a keen interest in G4s and their binding transcription factors (TF) as druggable targets. It is hypothesised that by disrupting their interactions it will supress the corresponding gene and provide an effective treatment for cancer. Conventionally this has been achieved using planar aromatic molecules that bind an exposed G4 tetrad or intercalate through π-π stacking interactions and prevents binding of the TF. Though this method is undoubtedly effective there is an important issue of selectivity in that the molecule will bind multiple types of G4 causing unwanted off-target effects and as result none of these molecules have reached clinical trials.
An alternative method to this is the so-called “decoy strategy” whereby an oligonucleotide which has the same or similar sequence to the wild type. This oligo will fold into a G4 structure and sequester the TF in direct competition to the endogenous G4 preventing transcription. This method shows great potential for selective interactions with TFs however there are certain requirements for effectiveness. Namely, the oligonucleotide must adopt the correct topology for TF binding and must be nuclease resistant. This is achieved with extensive SAR studies using a variety of chemical modifications such as LNAs at flanking regions and covalently bond polycyclic aromatics.
The aim of this project is therefore to develop new simplified G4 decoy stabilisation techniques leveraging the Madder group’s experience in DNA crosslinking moieties. The outcomes of this will be two-fold, firstly creating more stable and effective decoys and secondly, mechanistically we will be able to the explore the importance of topological interconversion for the binding of G4s to proteins.